DOI: 10.1093/molbev/msz185
- 1. School of Biological and Chemical Sciences, Queen Mary University
- 2. School of Chemistry and Molecular Biosciences, University
- 3. Department of Biotechnology, Indian Institute of Technology Kharagpur
- 4. Department of Computer Science, Royal Holloway University
- 5. Scientific Computing Facility, Max Planck Institute of Molecular Cell Biology and Genetics
- 6. San Francisco, CA
- 7. Computational Bioscience Research Center
- 8. King Abdullah University of Science and Technology
- 9. Bioinformatics Hub, School of Biological Sciences, University
- 10. Institute of Evolutionary Biology, University
- 11. Spiber Inc.
- 12. Whitehead Institute for Biomedical Research
- 13. Living Systems Institute, University of Exeter, Exeter, United Kingdom
- 14. The Institute for Genomic Medicine
- 15. The Abigail Wexner Research Institute at Nationwide Children’s Hospital, Columbus,
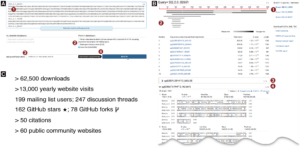
From “Sequenceserver: A Modern Graphical User Interface for Custom BLAST Databases” by Anurag Priyam et al.. Licensed under CC-BY 4.0
Abstract
Comparing newly obtained and previously known nucleotide and amino-acid sequences underpins modern biological research. BLAST is a well-established tool for such comparisons but is challenging to use on new data sets. We combined a user-centric design philosophy with sustainable software development approaches to create Sequenceserver, a tool for running BLAST and visually inspecting BLAST results for biological interpretation. Sequenceserver uses simple algorithms to prevent potential analysis errors and provides flexible text-based and visual outputs to support researcher productivity. Our software can be rapidly installed for use by individuals or on shared servers.